Quick start
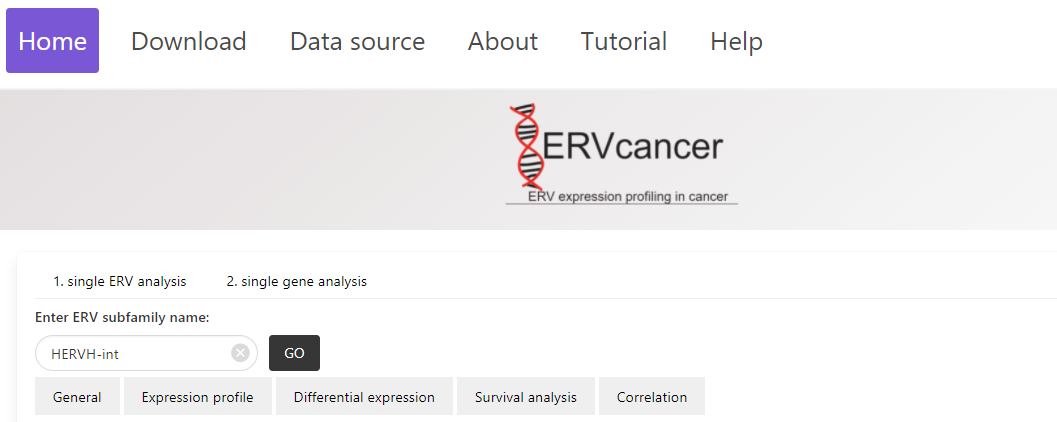
1. Enter an ERV name in the “Enter ERV subfamily name” field of “1.single ERV analysis”, and click the “Go” button to search the ERV of interest.
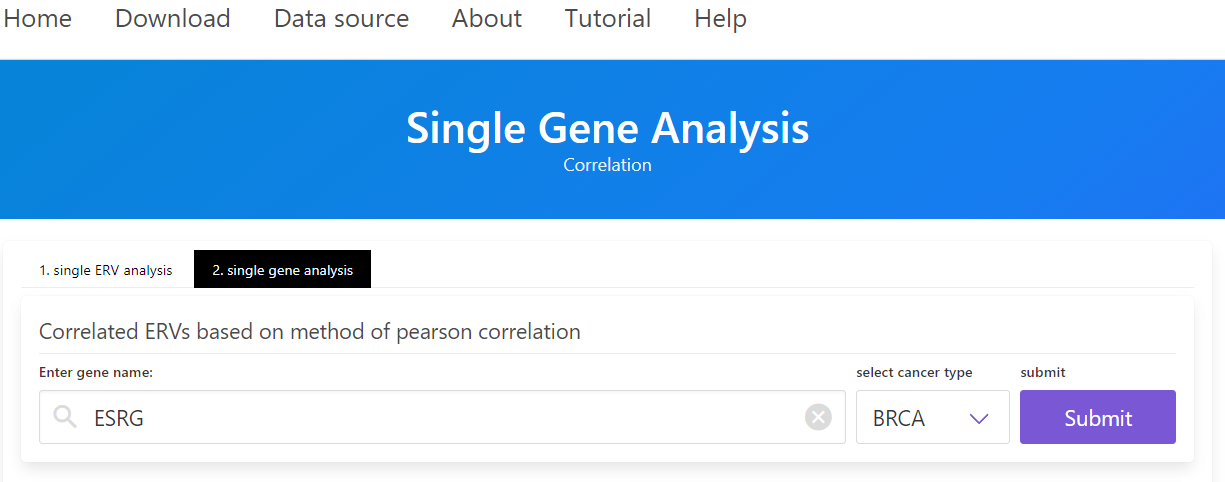
2. Enter a gene name in the “Enter gene name” field of “2.singe gene analysis”, select the cancer type, and then click the “Go” button to search the gene of interest.
Features in ERVcancer
1. Expression profile
(1) Parameters:
*Select samples: select a sample type of interest.*Select stages/tissue/cancer: select a stage or tissue or cancer type of interest.
(2) Results:
Click the submit button, ERVcancer will show expression profile based on input parameters.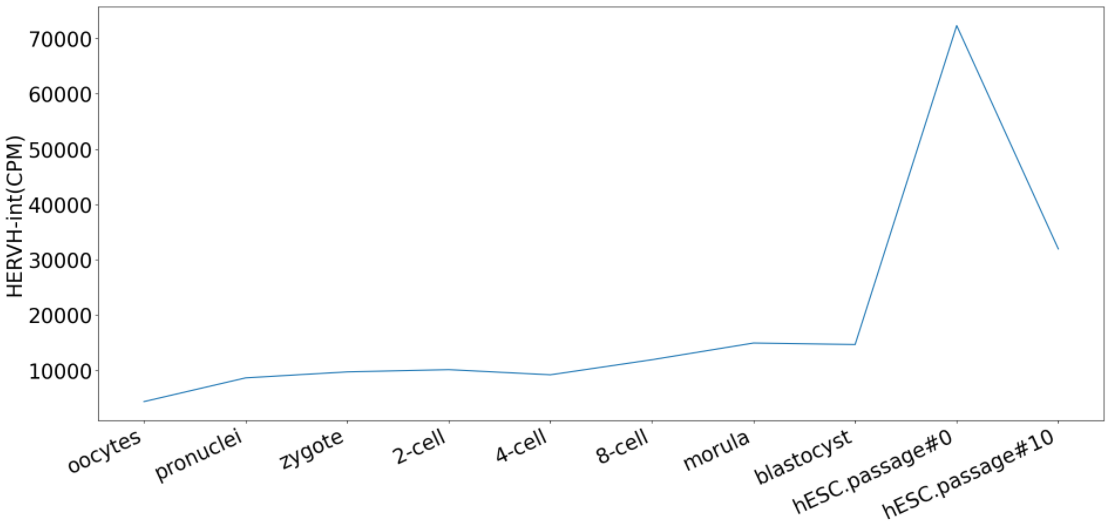
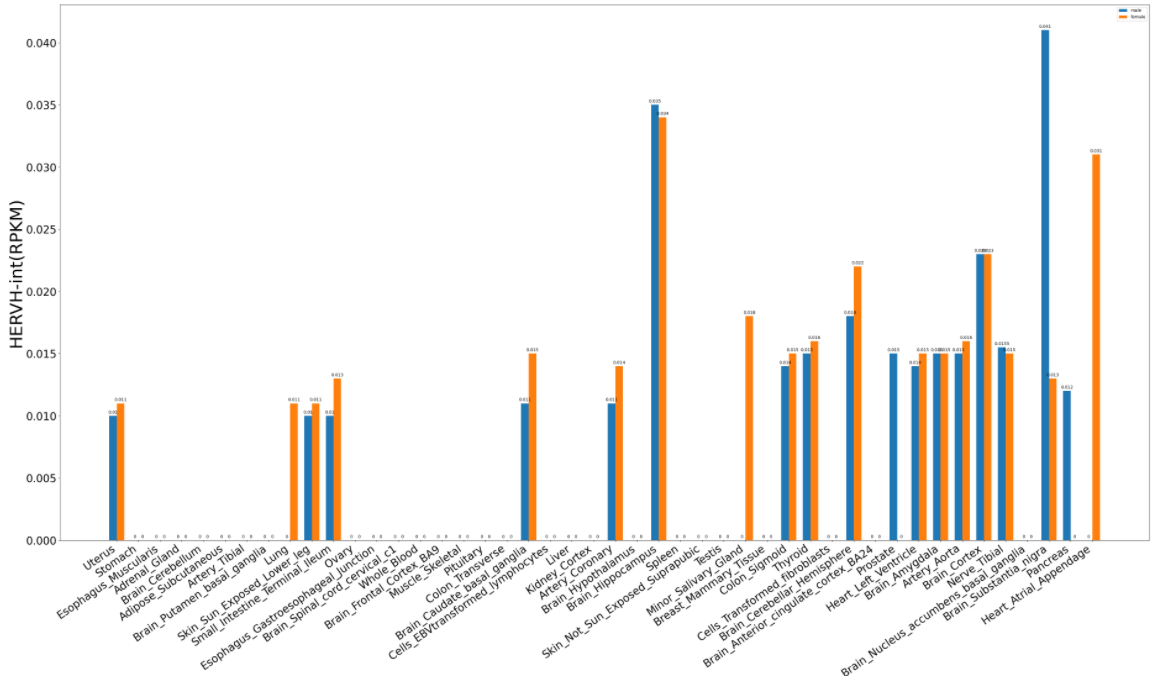
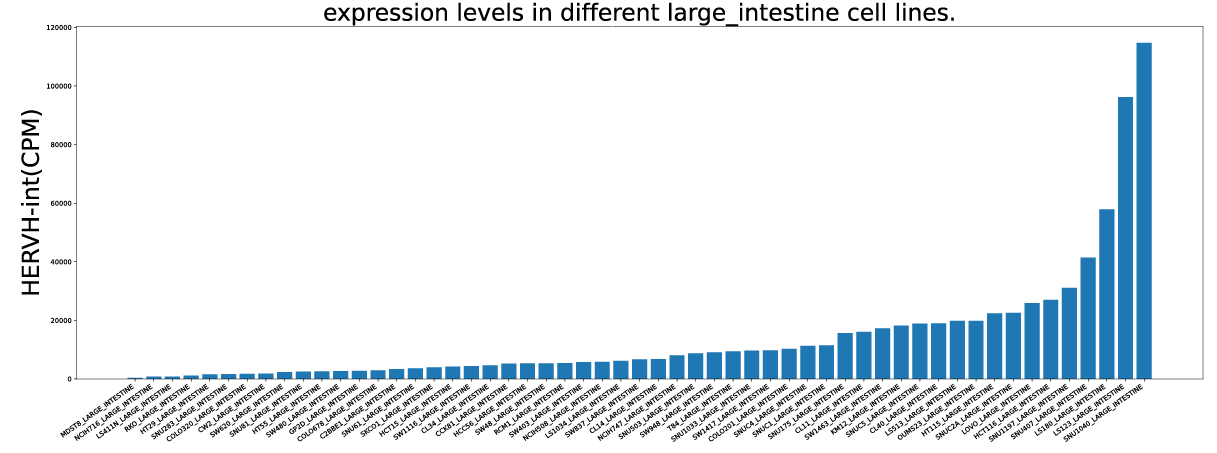
2. Differential expression
(1) Parameters:
*Select a cancer type: select a sample type of interest.*Select a condition for comparison: select a condition for comparison of interest.
*Method: select a statistical method to compare normal and tumor tissues expression level (CPM), or CPM among different stages or histological types.
(2) Results:
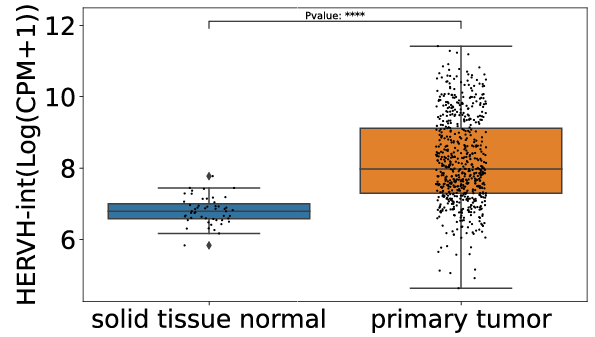
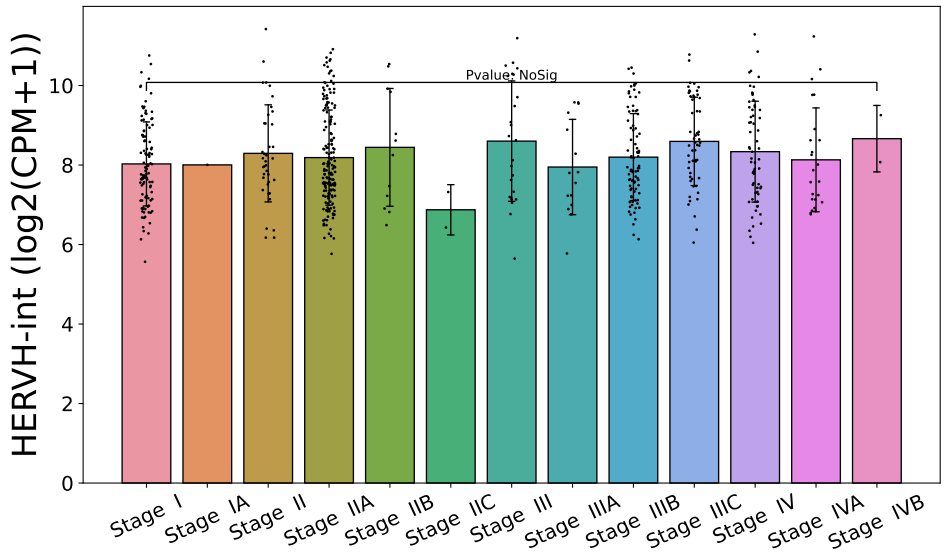
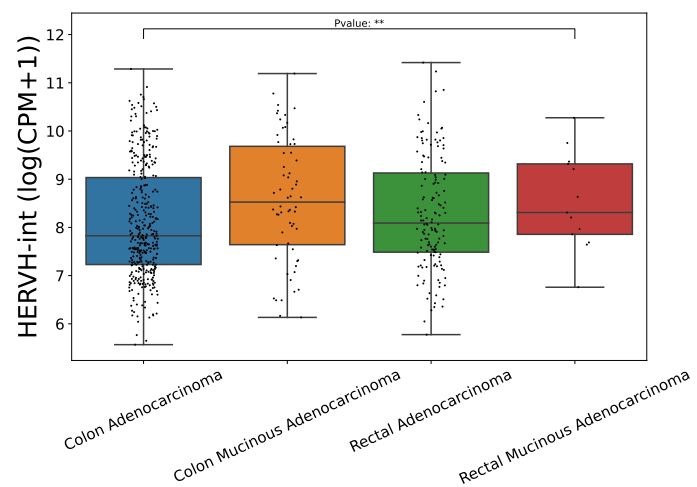
Click the submit button, ERVcancer will showa. differential expression result based on comparison of cancer and normal tissues and user’s custom input parameters

b. tumor or normal tissue sample CPM of specific ERV
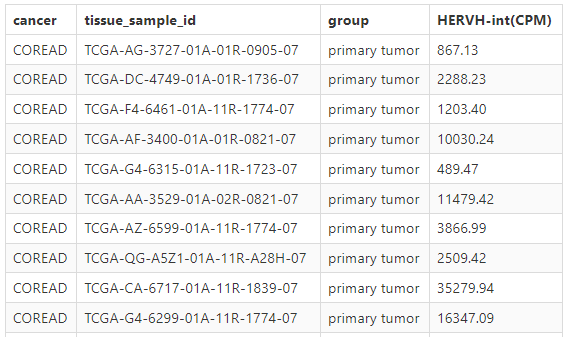
c. box plot or bar plot of specific ERV in chosen cancer type based on chosen method. Each dot represents a distinct tissue sample.
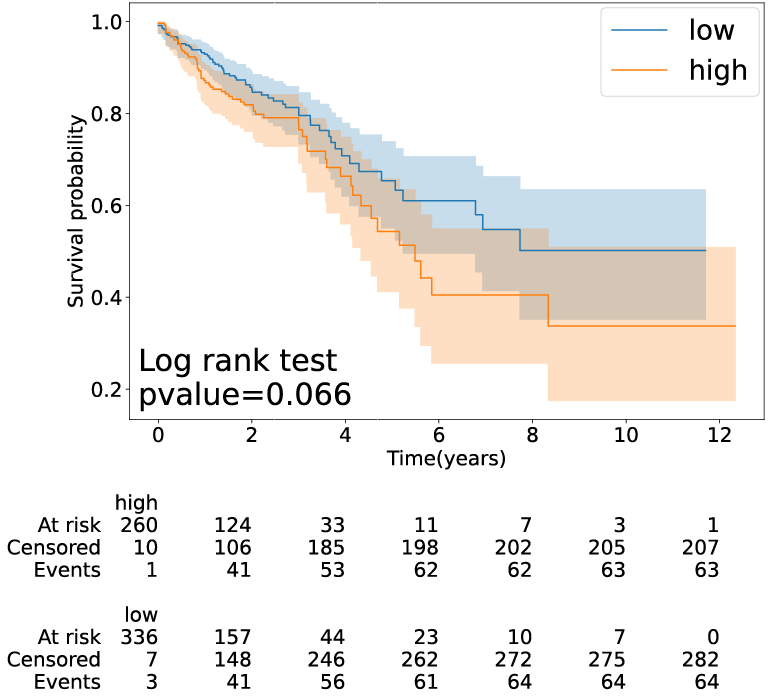
3. Survival analysis
(1) Parameters:
*Select a cancer type: select a cancer type of interest*Methods: select a method to classify patients into different groups.
(2) Results:
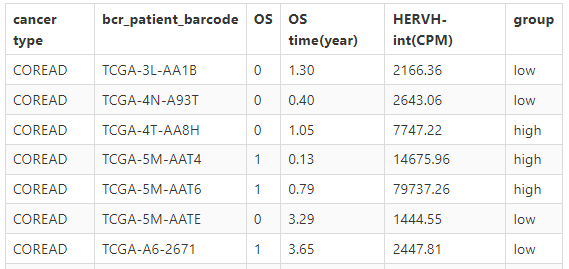
Click the submit button, ERVcancer will showa. Patients ERV CPM and their OS and OS time information
b. Survival analysis result
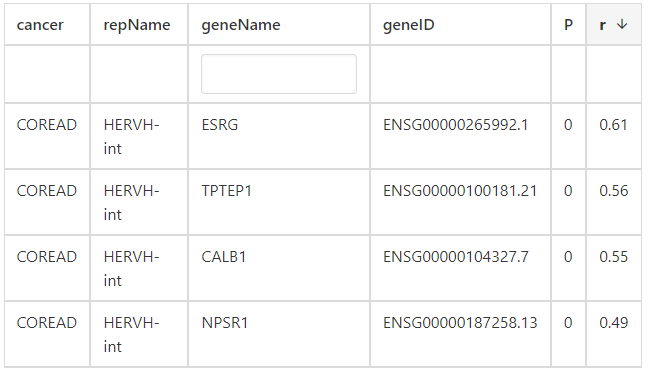
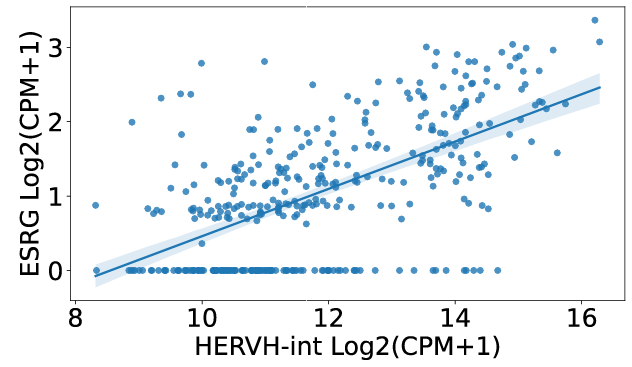
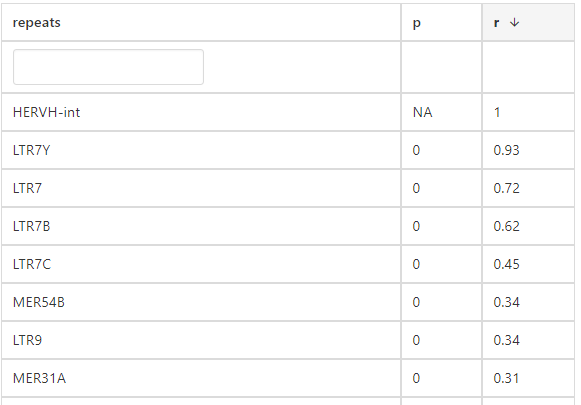
4. Correlation
(1) Parameters:
*Select a cancer type: select a cancer type of interest*Method: select results of interest
(2) Results:
click the submit button, ERVcancer will show correlation result based on Pearson method:a. Correlated ERVs
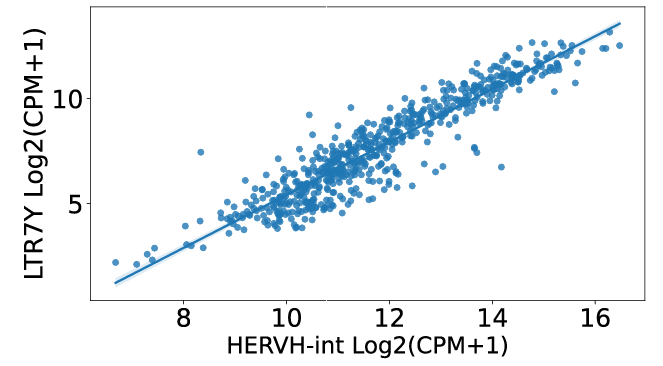
b. correlated genes