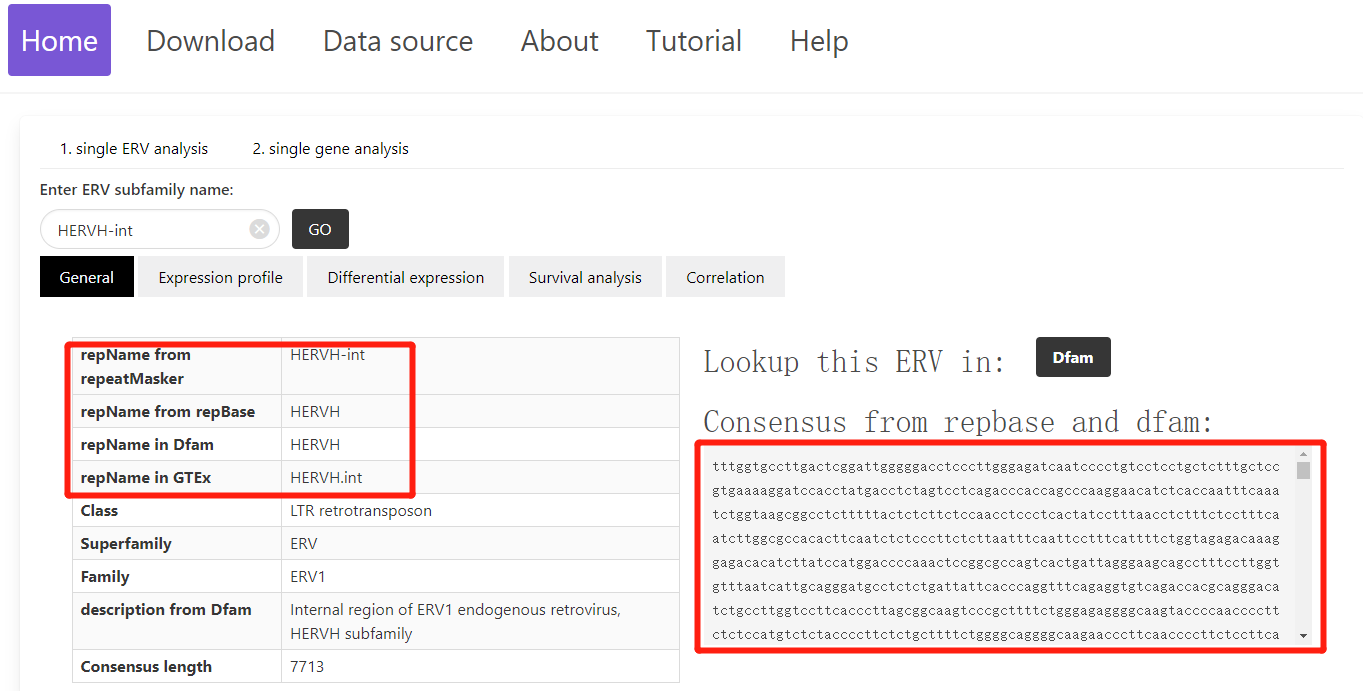
1. General
Using ERVcancer, experimental biologists can easily explore the extensive TCGA and CCLE datasets, pose specific research questions, and test their hypotheses. In the "General" section of the single ERV analysis, users can access a comprehensive overview of a specific ERV, including its name across various tools, websites, or datasets, as well as its consensus sequence.
2. Expression profile
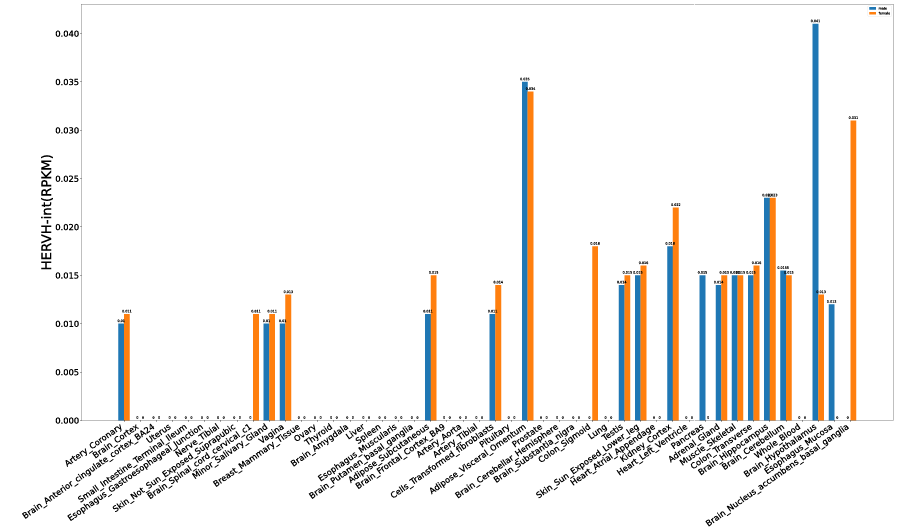
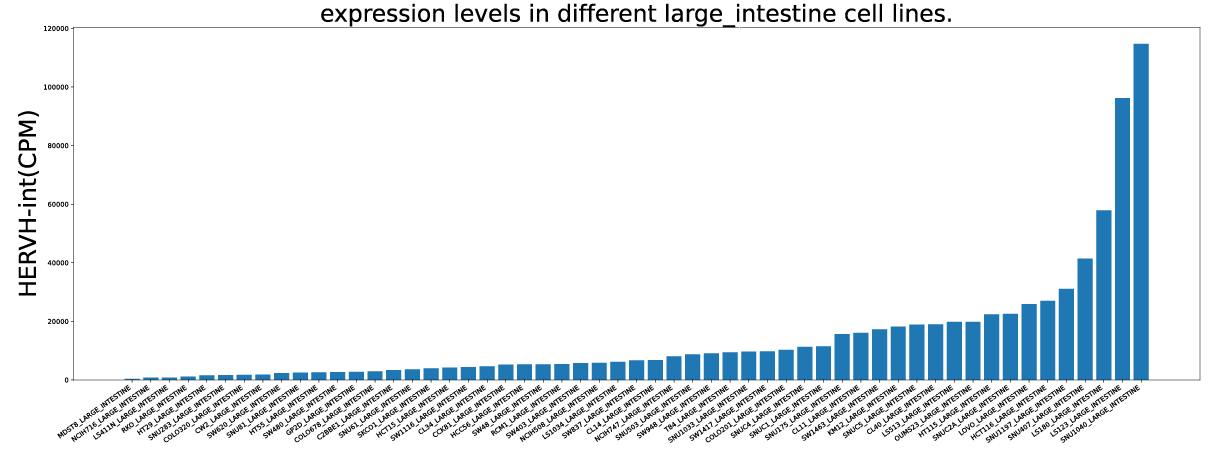
Additionally, based on the expression profiles, users can explore ERV expression levels during early embryogenesis, in adult human tissues, and across cancer cell lines.

3. Differential expression
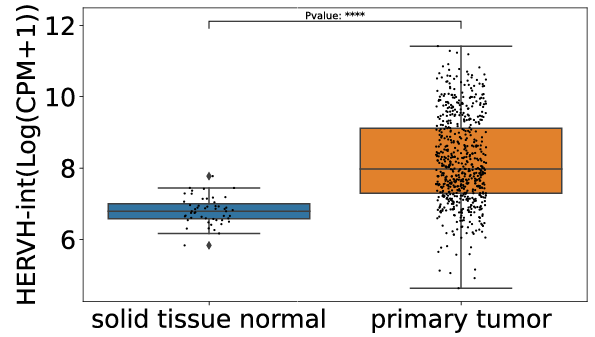
Users can utilize differential expression analysis to discover that HERVH-int is overexpressed in COREAD cancer compared to normal tissues.
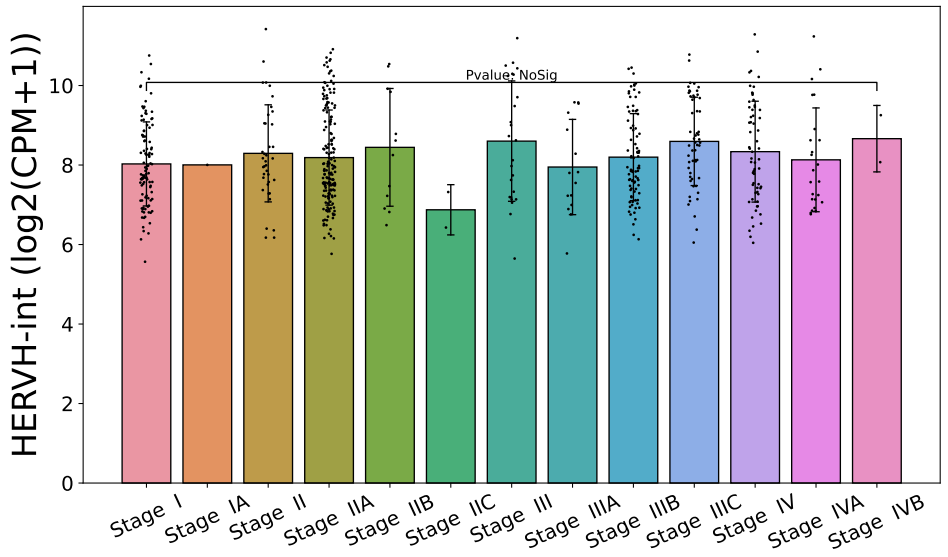
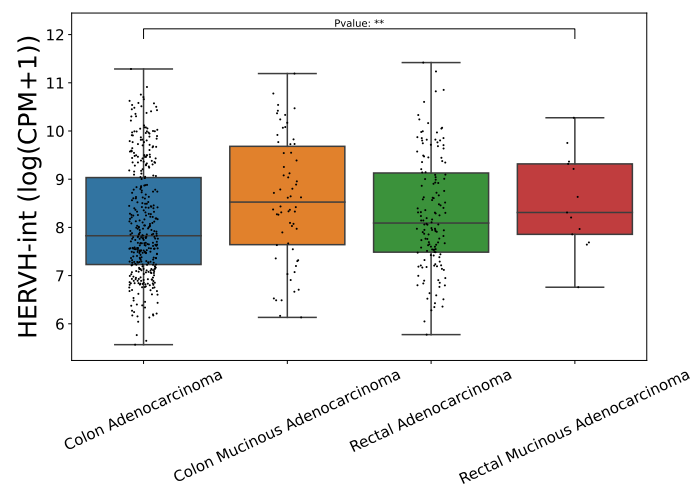
There is no significant difference in expression across different stages of the disease, but mucinous adenocarcinoma shows a higher expression level of HERVH-int.
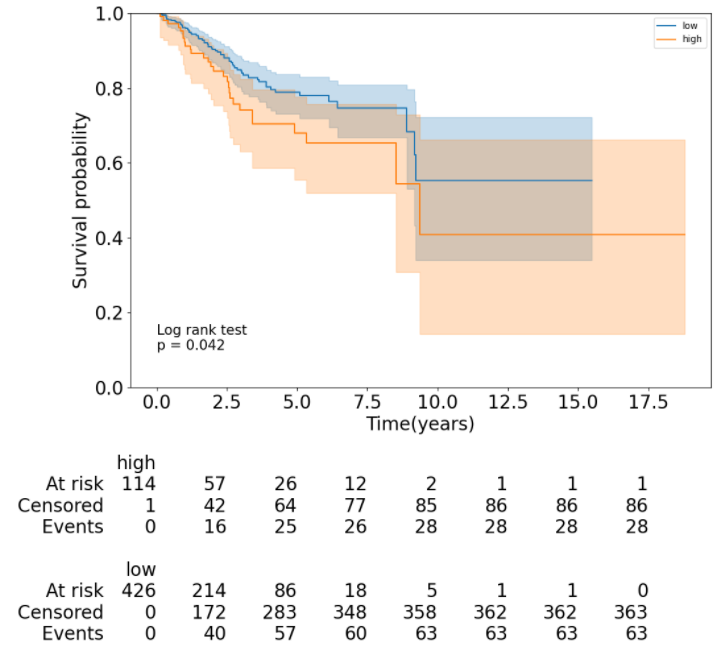
4. Survival analysis
For survival analysis, users can apply different methods to classify patients into distinct groups and compare their clinical outcomes.
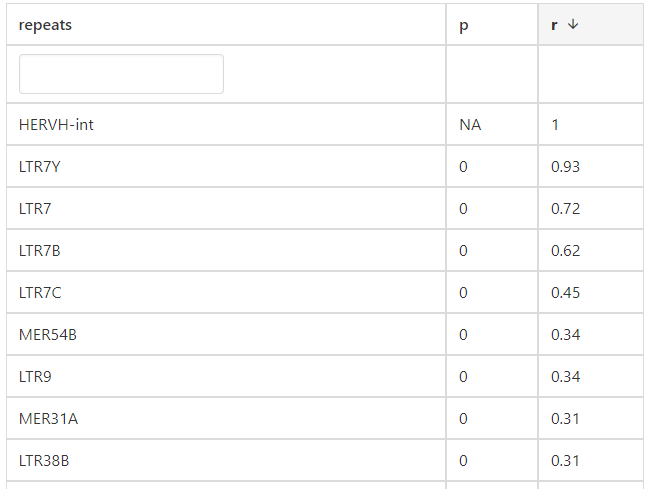
5. Correlation
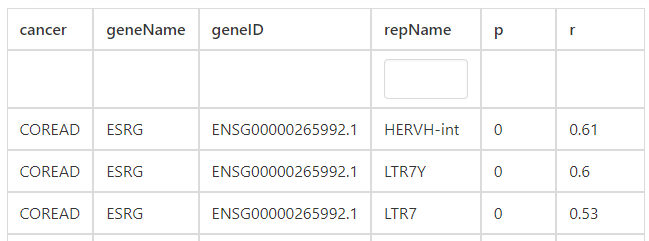
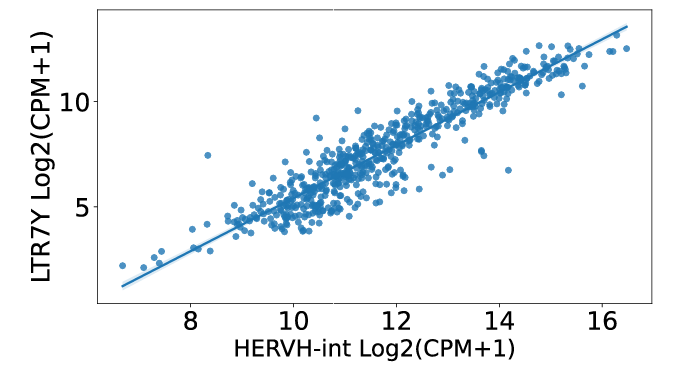
In the correlation section, users can identify that LTR7Y, LTR7, LTR7B, and LTR7C exhibit the highest correlation with HERVH-int in COREAD cancer.
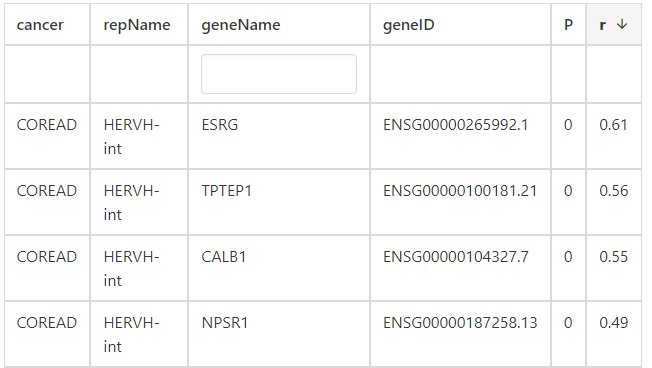
For correlated genes of correlation section, users can find that ESRG, TPTEP1, CALB1 and NPSR1 have the highest correlation with HERVH-int in COREAD cancer type.
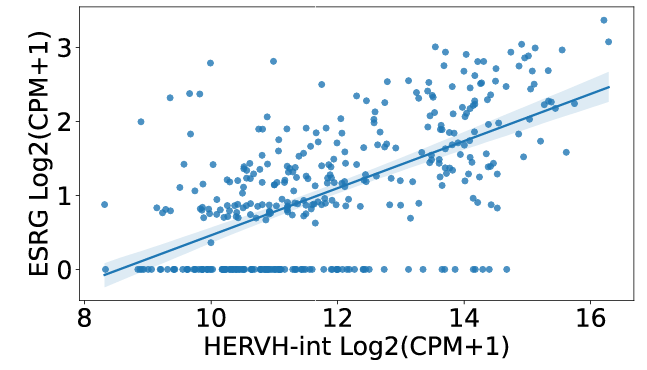
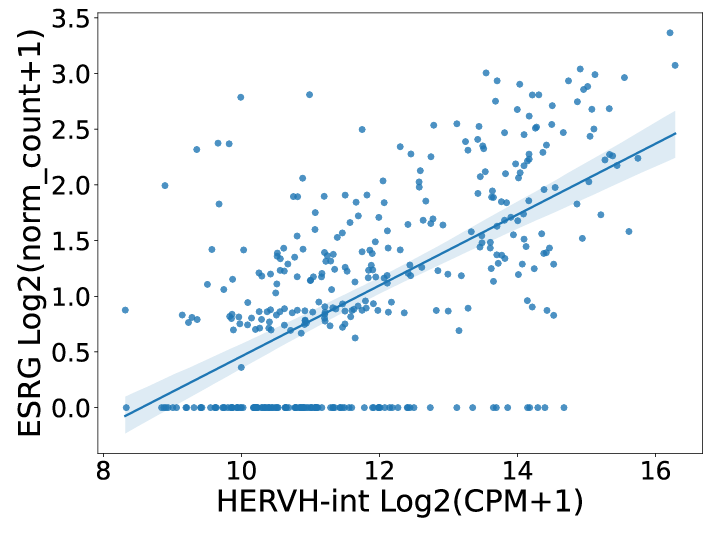
For single gene analysis, users can find correlated ERVs based on Pearson correlation method to find similar ERVs of specific gene. For example, users can find that HERVH-int, LTR7Y, and LTR7 have the highest correlation with ESRG in COREAD cancer type.